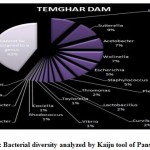
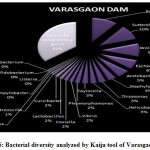
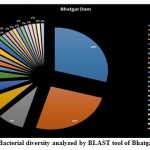
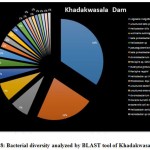
Introduction
Soil is a dynamic entity and a good platform for the exchange of matter and energy. It provides the main niche for all kinds of microorganisms including fungi (90% of biomass of soil is composed of bacteria and fungi)1. Mycotoxins produced by some fungi like Fusarium not only spoil farm yield but also harm the consumers. Hence, antimycotics are used over crop plants to prevent post-harvest loss that indirectly also effect human health. The last five decades has heralded the introduction of azoles as a popular fungicide in agriculture and clinics. Azoles such as dithiocarbamates, griseofulvin, ciclopirox, and strobilurins are commonly used as antimycotics2. Lately, it has been observed that overuse of azoles as fungicides in agriculture can lead to cross-resistance among various fungal species and is the main cause of emergence of novel resistant fungi such as Candida aurisin clinical samples3.
Human fungal infections generally do not receive much attention as compared to bacterial and viral diseases; however, mortality rate from invasive fungal infections is increasing exponentially despite the use of antifungal drugs4. Among the common fungal infections afflicting human, candidiasis affects millions of people annually. Mainly associated with hospitalization, the infection usually spares the vast majority of immunocompetent patients but has a predilection for immunocompromised individuals5. It has been reflected as the predominant systemic infection causing pathogen in patients with COPD, AIDS, and cancer6,7,8,9,10,11. Most importantly, the resistant isolates of Candida have the potential to form biofilms. The thick polymeric extracellular layer protects Candida aggregates as it forms a tough barrier against the known antimicrobial substances12.
The emergence of clinical Candida isolates with multi drug resistance (MDR) has further alarmed microbiologists. The usual Candida species predominantly found in clinics are susceptible to amphotericin B, azoles, and echinocandins, but certain novel emerging fungal species have demonstrated resistance to all the three primary antifungal drugs13. Ironically, amphotericin B performs slightly better whereas azoles have confirmed to be the least effective against such species14. Though in most clinical isolates, resistance to antifungal is due to long term azole therapy, wherein the fungus was adapting rapidly to the azole drug being used; however it was also seen that certain patients who were never exposed to azole drugs, showed resistance. Such incidences are a wakeup call and indicate that these patients perhaps acquired azole resistant fungal strains from the environment.
Candida species in patients andenvironment
Candida is an important opportunistic fungal pathogen that is present both in immunocompromised patients and also in the environment. It causes infections in mouth, skin, and vagina along with bloodstream invasive infections. Candida albicans is the major causative agent of all forms of candidiasis. However, the pattern has changed in the last decade with majority of the non albicans Candida species (NACS) getting associated with the nosocomial invasive infections.C. glabrata, C. parapsilosisand C. tropicalisare the important pathogens in this category. Higher mortality rate in patients with C. glabrata over C. albicans is due to the fact that it can survive during long term starvation conditions upon phagocytosis by macrophages, as well as low pH environment in vagina or in phagolysosomes. C. glabrata thus is associated with the enormous adaptability to survive in different host niches. C. tropicalis is a diploid dimorphic fungal pathogen which causes infections mainly in neutropenic patients and individuals having haematological malignancies15. Similarly, C. parapsilosisis also a diploid dimorphic fungus which mainly affects the immunocompromised individuals and can cause major health risk to neonates with low weight birth.
Almost ubiquitously, many Candida species are found in every kind of managed soil (Table 1); however variation depends on type of soil, climate condition, water abundance etc1. Fungi ferment and assimilate different kind of sugars provided by the plants to soil e.g.C. auris ferment glucose, sucrose but assimilate maltose, mannitol, sorbitol as carbon sources16,17. The abundance of different sugars in different soil decides the variation in fungal community which further gets enhanced due to regular tillage and mixing of upper layer of soil.
Table 1: Candida species isolated from different kinds of managed soil
| Candida species present | Type of managed soil | References |
|
Candida glabrata |
Vineyard |
18 |
| Candida parapsilosis | Agricultural | 19 |
| Candida apis | Pasture | 20 |
| Candida azyma | Rhizosphere | 21 |
|
Candida diddensiae |
Rhizosphere |
21 |
| Candida sake | Agricultural |
22 |
| Candida (Torulopsis) magnolia | Agricultural | 19 |
Use of Azole in Agriculture and Medicine
Fungal infections in plants cause a massive loss in crop yield worldwide. In agriculture, antimycotics are mainly used to control the fungal growth on plantsand also to arrest post-harvest spoilage of agricultural product. Though before 1970s, many compounds such as dithiocarbamate, benzimidazoles, strobilurins were predominantly used antimyocotics; however later azoles gained prominence due to their site-specific inhibitory action which made them much more beneficial2. According to the instructions of manufacturers, it is stated that about 10mg of azole should be applied on 1m2 plant surfaces, or in other words, about 100g/ha should be used in a field23. As per the available data, usage of azole-based fungicide has increased from 100 tons to 145 tons i.e. 45% in 4 years in Taiwan (2005 to 2009)24. Throughout the world, fungicide share 27% of anti-pest chemicals and this number is 15% in India; which is rising exponentially to the demand of increasing population25. Certain features of azoles make them preferred fungicides among farmers as they possess long–lasting stability and high efficiency against broad range of plant infections. The azole fungicides are selective only for fungi species and target the ergosterol biosynthetic pathways in the fungus. They also show very low toxicity for both the farmers who apply them and the plants that are under treatment, as both plant and animal cells do not contain ergosterol in their plasma membrane which is an important component of fungal cell membrane.The massive use of azoles in agriculture however, exerts a strong selective pressure on the pathogenic fungi to survive in environment.
In medicine on the other hand, the increase in prophylactic usage of therapeutic azoles has led to surge in the cases of resistant fungal isolates. Previously C. albicans was considered as a ubiquitous fungus in hospitals and cause of nosocomial infections26,27,28; but nowadays, other species have also paved their way to the hospital wards. These non-albicans Candida species (NACS) are distributed according to geographic areas e.g.C. tropicalis majorly found in Asia, in organic soil and soil near water29,30,31 while C. glabrata predominate in western countries 27, 32.
The similar mode of action of azoles used in agriculture and medicine perhaps has led to cross-resistance along with increased presence of resistant fungal strains in environment and in clinics.
Correlation between the use of azoles in agriculture and resistance in clinical fungi
Bacteria and fungus in soil develop antibiotic resistance mechanism as a natural process. However, the mechanism of resistance in fungi and its transmission differs significantly from that in bacteria. There is no evidence that genes that confers resistance in fungi are transferred horizontally, a major difference with bacteria33. Acquired resistance is less common than intrinsic resistance in fungi because fungi do not exchange the resistance genes via plasmids, therefore selection of resistance mechanisms in fungi occur as an outcome of the antifungal use34. Most of the pathogenic fungi have their natural habitat in the environment and spores of fungi generally spread over long distance by air flow. Many fungal pathogens are present in both environment and clinicse .g. Aspergillusfumigatus, a saprophytic fungi that survive on decaying organic substances like leaves, fruits, etc. These fungi produce spores that are blown off by air and spread to other organisms e.g. humans. These spores mainly cause Aspergillosis in humans. As inhalation is the main route of these spores, so if an azole-resistant A. fumigatus is present in human surroundings then this resistant strain can enter the human body and can lead to the development of azole-resistant fungal disease35. Notwithstanding, compost present in gardens and in indoor plants could also act as an ecological niche for the various spores of fungi as well as azole residues; and can play an important role in development of azole resistance36.
Clinical azole drugs represent the major core therapy for treating fungal diseases in human35. Normally, treatment of the patients over longer period of time with antifungals can result in the selection of resistant strains. However it has been seen that exposure of fungi to azoles in agricultural field develops cross resistance to medical triazoles37. Due to the presence of genetic homology between the clinical and agricultural isolates, it has been suggested that environment is the most common infection route for the emergence of azole resistant isolates in clinics and hospitals. In a study, it was found out that 50% isolates of C. tropicalis isolated from the soil are 97% identical to those of human C. tropicalis while 33.3% are completely identical (100%) to human C. tropicalis. It was also seen that C. tropicalis isolated from humans and soil with reduced susceptibility to fluconazole are showing reduced susceptibility to other azoles that are in continuous use in agriculture31.
Even though development of resistance is a complex process and takes time to achieve, extensive data has revealed that azoles lead to strong selective pressure over fungal population to choose resistant strains e.g. Benzimidazole when initially introduced was a revolutionary agent against pathogen but later on some strains of the pathogen which are naturally resistant to benzimidazole got selected naturally and emerged further38. This forms a major concern for microbiologists nowadays. Strains of fungi isolated from HIV patients pre-treated with azoles are found to be resistant to fluconazole and they also show cross-resistant to azoles used in agriculture, with high Minimum inhibitory concentration (MIC) values. Resistance to medically important antifungals imposes a big challenge to clinicians in treating invasive fungal diseases. The knowledge about the rate and extent of development of resistance mechanism in the fungi and whether the mutation associated with this resistance is reversible or not is a very important point in the clinical medicine. The resistance to variety of antifungal depends on many factors for instance, the pharmacokinetic and pharmacodynamics properties of the drug, the target site of the drug and its mode of action in organism. It has been studied that there are mainly three ways by which patients may acquire the azole resistant fungi (1) initially, the infecting strain of fungi is susceptible to antifungal but as it mutates it develops azole resistance, (2) the individual may harbour the heterogenous population of infecting strain due to which resistant strain get selected during antifungal treatment over a period of time, or (3) the individual may acquire resistant strain from the external milieu23.
Mode of action of azole antifungals in Candidasp.
Binding of azoles gets impaired by genetic alteration in ERG11gene which mainly accounts for more than 90% of azole resistance in clinical isolates. The ERG11 gene encodes the enzyme cytochrome P450 14α-demethylase (also called lanosterol 14α-demethylase) which is the primary target for the azole compounds. Azole compounds are classified into two types depending on the basis of nitrogen atom present in it. Imidazoles (miconazole, ketoconazole) contain two nitrogen atoms in the ring; while triazoles (fluconazole, itraconazole, posaconazole, voriconazole) have three nitrogen atoms in ring. The nitrogen atom plays an important role in the mechanism of action of azoles as it bind to the heme group of the enzyme encoded by the gene. The enzyme encoded by the ERG11 plays a major role in removing the methyl group from lanosterol and then converting it into ergosterol. The ergosterol is a major component of the plasma membrane of fungi; hence it has a major role in maintaining the integrity of the cell membrane and also participates in the cell signaling pathways. The azole compounds work by inhibiting the function of lanosterol 14α-demethylase enzyme which will result in the accumulation of the methylated toxic precursors (Fig. 1). The build-up of these precursors in the cell makes plasma membrane leaky and impairs the fungal growth.
 |
Figure 1: Action of azoles in Ergosterol biosynthetic pathway |
Mechanism of azole resistance in Candida sp.
Resistance to azole antifungals to Candida sp. has been extensively studied. In C. albicansazole resistance can be broadly attributed to (i) overexpression of ABC and MR1 efflux pumps ii) enhanced secretion of 14α-lanosteroldemethylasevia ERG11 gene up-regulation; iii) decreased affinity of azole drugs due to point mutations in ERG11 gene and iv) homozygous ERG3 deletion mutation leading to the loss of sterol 5,6-desaturase activity39,40,41. These mechanisms are elucidated as under
Overexpression of Efflux Pump
Mutations in trans-regulatory factors culminate into up regulation of genes encoding for efflux pumps and are the major cause for drug-resistance in C.albicans isolates(Fig. 2).ATP-binding cassette (ABC) is the primary active transporter of the cell membrane having four domains- two Nucleotide-binding domain (NBD1 and NBD2) that binds to ATP and hydrolyse it to let the solute into the cell and the other two domains are the transmembrane domain (TMD1 and TMD2)42. In C. albicans for azole resistance, mainly ABC transporter comes in focus as out of two ABC transporter CDR1 and CDR2, CDR1 plays a major role and get upregulated43,44. In C. auris, along with CDR1 and CDR2, another ABC transporter Snq2 plays a role in azole resistance. Various transcriptomic studies have concluded that efflux pump′s over expression along with some zinc cluster transcriptional factors e.g. TAC1 (transcriptional activator of CDR genes) are a cause of azole resistance. Exposure to fluconazole, cause mutations in TAC1B which is located near the mating type locus, as the major regulator of CDR1 and CDR2 and this was found in many isolates of flucon azole-resistant C. auris45. Over expression of another efflux pump, MDR1 has also been found to be responsible for drug resistance in C.albicans isolates. Genetic alterations in MRR1 (multidrug resistance regulator 1), a zinc cluster transcription factor is the major factor for MDR1 over expression and multidrug resistance. Mrr1p target genes encode oxido reductases and it has been suggested that MRR1 gain-of-function mutations may be in response to the oxidative damage of the cells due to azoles, and the up regulation of Mrr1p target genes may play a role to restore the intra cellular redox balance; thereby causing resistance46.
 |
Figure 2: Overexpression of (a) ABC and (b) MDR1 efflux pump in azole-resistant Candida sp. |
Increased expression of ERG11 gene and enhanced ergosterol synthesis
Expression of a target enzyme is also up regulated in cell by increased transcription rate of a gene; by gene amplification; and by decreased degradation of the gene product. Additionally, transcriptional factors may play a major role in expression of gene and susceptibility towards azole drug. The Upc2p is a zinc finger transcription factor and acts as a key regulator that regulates the expression of genes involved in ergosterol biosynthesis, including ERG11 gene47. In C. albicans, a gain of functional mutation of Upc2p may lead to the increased production of the ergosterol, hence leading to resistance to fluconazole whereas, disruption of Upc2p results in enhanced fluconazole (FLC) activity. It was hence observed that Upc2p affects the azole susceptibility viaergosterol biosynthesis48,49.
Point mutations in ERG11 gene and amino acid substitution
The point mutations (insertion and deletion) in the ERG11 gene leads to the reduced binding of the azole drug and results in the origin of the clinically resistant isolates (Fig. 3). Mechanism of resistance is mostly studied in C. albicans. Many amino acid substitutions have been found in the ERG11 gene product but only a few are experimentally confirmed to have a link with azole resistance50. Usually, amino acid substitution occurs in the hot spot regions of 105-165, 266-287, 405-488bpin Erg11p40. The molecular analysis has revealed that these substitutions occur in the catalytic site of the enzyme due to which there is a decrease in binding affinity of drugs44,51. For C. auris some specific substitution is in focus e.g. in South American and South Asian clade of C. auris K143R and Y132F substitution takes place, while in African clade, F126L substitution is most common52. In the case of C.glabrata such substitutions were not found and only a single case of this kind of resistance mechanism was found in C. glabrata51.
 |
Figure 3: Mutation in Erg11 in azole-resistant Candida species |
Alteration of ergosterol biosynthetic pathway by inactivation of ERG-3 gene
Fungi is found in different kinds of environmental niches and is accompanied by diverse environmental stress53. For fungi, antifungals are also a kind of stress to which they adapt and try to survive43,54. The main mechanism of resistanceforC. albicans is the remodeling of the ergosterol biosynthesis pathway. This occurs as a result of the loss of functional mutation in ERG-3gene. ERG-3 gene is responsible for coding Δ-5,6desaturase and because of loss of function mutation, deactivation of this enzyme occurs due to which there is no accumulation of methylated sterols formed due to azole action on the fungal membrane55. As a result there will be less loss of fungal cell by azoles i.e. decreased susceptibility of fungi to azoles. In C. albicans there are five missense mutations and two non-sense mutations, which are the reason for the loss of function and resistance for azoles50,56.
As true for stress response in every organism, chaperone such as (Hsp 90) works well in the fungal cell also. Hsp90 is a protein folding molecule and is considered as the main molecule involved in developing resistance in C. albicans. Alternatively, it was also observed that a very high amount of doxycycline is required to inhibit Hsp90 function in C. auris than C. albicans57. Hence, in C. auris, azole resistance arises due to increase in the function of efflux pump and is observed to be independent of Hsp90 57,58.
Besides the above important mechanisms, aneuploidy has also been found to play a role in azole resistance in Candida.Aneuploidy leads to genetic diversity by allelic imbalances and loss of heterozygosity (LOH) at any of parental allele. In fungal pathogen, this aneuploidy and LOH results in a change of working pattern of efflux pump and change in action site of the drug; and these factors together lead to resistance in fungi. Usually, a fungal cell has a good genetic diversity which results in phenotypic plasticity and ultimately resistance. In C. albicans, by the different combination of hybridization, it was found out that aneuploidy occurs on chromosome 1, 2, 5 which usually contributes to the resistance initiation59,60. This LOH often leads to the gain of function mutation for the efflux pump and other drug targets resulting in resistance. In C. albicans aneuploidy on chromosome 5 is more common and this resistance is linked to a high copy number of ERG11 and TAC1 gene60,61.
Modes of transmission of resistant fungi
The environmental fungi undergo transmission from environment to hospital wards and exhibits cross-resistance to clinical azoles. Air can be a possible source for transmission of fungi from field to hospital especially the transmission of those fungi that sporulatee.g. A.fumigatus. Insects (especially insects belonging to Muscidae, Calliphoridae class) can also act as a means of transmission for resistant fungus62,63,64. Direct contact of farmers and workers during field management and post-harvest processing may also transfer fungus from field to humans. Raw fruits and vegetables are considered as a good source of nutrients and part ofgood healthy diet but their associated fungi have become a point of concern because of increasing resistance to antifungals. There is an increasing demand for these fresh raw fruits and vegetables that led to 25% increase in consumption of fruits from 1977-1979 to 1997-1999 in the USA65. This had prompted some industries to conduct risk analysis and management practice for these fresh produces but still, there are increasing number of diseases associated with fruit-consumption66. Researchers working in this field face the major challenge of lack of information among affected patients on the source of contamination and procurement of the raw fruits and vegetables consumed67. As all of these products usually follow a chain system in which farmer provide these resources to the wholesale market; and from these markets different distribution agency procures goods; so ultimately no one knows the real source of the infected fruits and vegetables. In 1995, there was a simultaneous breakout of SalmonellaStanley due to usage of alfalfa sprouts in the USA and Finland; which was finally traced down to a common source of contaminated seeds shipped by a merchant68. In different studies, it was found out that raw fruits and vegetables are also a good source for microbial transmission from field to human. In a study, nearly 184 yeasts were isolated that had a high percentage of Candida species responsible for causing diseases in human and all of these species showed decreased susceptibility to fluconazole (MIC ≥8mg/ml)24. Another study conducted in 14 countries over a population of 883 million; showed that nearly 2% population in every country is infected with fungal infection69. These results hence suggest the path for transmission of resistant fungi from the field to humans. Different agencies and institutes e.g. Food and drug administration (FDA), European medicines agency (EMA) regulate the usage of antifungal in fields70. CLSI (Clinical and Laboratory Standards Institute) and EUCAST (European Committee on Antimicrobial Susceptibility Testing) have provided protocol to check the susceptibility of different fungi for different drugs3. But there is no such committee to check and regulate the usage of azoles in different fields in India. Hence there is an urgent need to make guidelines for usage of azoles in agriculture that could break this chain of transmission of resistant fungal strains from fields to human.
Conclusion
The fungal strains that are resistant to azoles have a great impact on human health. They generally acquire the resistance mechanism in the form of efflux pumps that lowers the concentration of a drug inside the cell or by other genetic modifications that decreases the susceptibility to drugs. The increase in azole resistance eventually leads to increase in virulence of such strains. Infections caused by these resistant isolates are more difficult to control by host defense mechanisms. Development of resistance to antifungal drugs generally leads to treatment failure in patients. However therapeutic failure in patients is dependent on many factors like immune status of host, site and severity of infection, drug interaction with target site, dosage of drug, cellularorganisation of the fungi, hyphal morphology, serotype, biofilm production and fungal load.Research in this field should focus on to establish the inter-linkages between the mutant fungal strains, theirresistance to agricultural azoles and eventual cross resistance to medical triazoles. However, even if relationship between the use of azole compounds and development of resistance in Candida is proven, further control of its spread and development of resistant isolates is almost impossible to curtail as they are already present in environment. The only limiting factor should be the judicial use of fungicides in horticulture and agriculture and stringent regulatory practices and surveillance for clinical testing of cultures of fungal isolates in human patients.
Acknowledgement
The authors express gratitude to Principal, Ramjas College, University of Delhi, for constant encouragement. KW, KJ and AY also wish to acknowledge Council of Scientific and Industrial Research (CSIR) and University Grants Commission (UGC) for financial support in the form of Junior Research fellowships.
Conflict of Interest
The authors declare that there are no competing interests.
References
- Nielsen M.N., Winding A.Microorganisms as indicators of soil health. National Environmental Research Institute. NERI Technical Report No. 388, Denmark 2002
- Russel P.E. A century of fungicide evolution. Journal of Agricultural Science 2005;143:11–25.
CrossRef - Ribas E., Ribas A.D., Spolti P., Del Ponte E.M., Donato K.Z., Schrekker H.,Fuentefria A.M.Is the emergence of fungal resistance to medical triazoles related to their use in the agroecosystems? A mini review. Brazilian journal of microbiology: publication of the Brazilian Society for Microbiology2016; 47(4): 793–799.
CrossRef - Fisher M.C., Henk D.A., Briggs C.J., Brownstein J.S., Madoff L.C., McCraw S.L.,Gurr S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature2012; 484(7393): 186-194.
CrossRef - , Ferrari S., Coste A.T. Antifungal resistance and new strategies to control fungal infections. International journal of microbiology 2012; 2012: 713687.
CrossRef - Marr K.A., Carter R.A., Boeckh M., Martin P., Corey L. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood2002; 100(13): 4358–4366.
CrossRef - Guinea J., Torres-Narbona M., Gijón P., Muñoz P., Pozo F., Peláez T., de Miguel J.,Bouza, E. Pulmonary aspergillosis in patients with chronic obstructive pulmonary disease: incidence, risk factors, and outcome. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases2010; 16(7): 870–877.
CrossRef - Brown G.D., Denning D.W., Gow N.A., Levitz S.M., Netea M.G., White T.C. Hidden killers: human fungal infections. Science translational medicine2012; 4(165): 165rv13.
CrossRef - Armstrong-James D., Meintjes G., Brown G.D. A neglected epidemic: fungal infections in HIV/AIDS. Trends in microbiology2014; 22(3), 120–127.
CrossRef - Denning D.W. Minimizing fungal disease deaths will allow the UNAIDS target of reducing annual AIDS deaths below 500 000 by 2020 to be realized. Philosophical transactions of the Royal Society of London. Series B, Biological Sciences2016; 371(1709): 20150468.
CrossRef - Limper A.H., Adenis A., Le T., Harrison T.S. Fungal infections in HIV/AIDS. The Lancet. Infectious diseases 2017; 17(11): e334–e343.
CrossRef - Cavalheiro M., Teixeira M.C. CandidaBiofilms: Threats, Challenges, and Promising strategies. Front Med (Lausanne)2018; 5:28.
CrossRef - Richtel M., Jacobs A.A.Mysterious Infection, Spanning the Globe in a Climate of Secrecy. New York Times. from https://www.nytimes.com/2019/04/06/health/drug-resistant-candida-auris.html.2019.
- Kaufmann C.A., Marr K.A.,Thorner A.R. Management of candidemia and invasive candidiasis in adults. UpToDate. from https://www.uptodate.com/contents/management-of-candidemia-and-invasive-candidiasis-in-adults. 2019.
- Whaley S.G., Berkow E.L., Rybak J.M., Nishimoto A.T., Barker K.S., Rogers P.D. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Frontiers in microbiology2017; 7: 2173.
CrossRef - OseiSekyere J. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. MicrobiologyOpen2018; 7(4), e00578.
CrossRef - Lone S.A., Ahmad A. Candida auris-the growing menace to global health. Mycoses2019; 62(8): 620–637.
CrossRef - Kachalkin A.V., Abdullabekova D.A., Magomedova E.S., Magomedov G.G., Chernov I.Y.Yeasts of the Vineyards in Dagestan and Other Regions. Mikrobiologiia2015; 84(3): 360-368.
CrossRef - Lund A. Studies on the ecology of yeasts. Munksgaard, Copenhagen.1954.
- Parle J.N., Di Menna M.E., The source of yeasts in New Zealand wines. New Zealand Journal of Agriculture Research1966; 9:98–107.
CrossRef - de Azeredo L.A., Gomes E.A., Mendonc a-Hagler L.C.,Hagler, A.N. Yeast communities associated with sugarcane in Campos, Rio de Janeiro, IntMicrobiol1998; 1:205–208.
- Wuczkowski M.,Prillinger H. Molecular identification of yeasts from soils of the alluvial forest national park along the river Danube downstream of Vienna, Austria (“Nationalpark Donauauen”). Microbiological research2004; 159(3): 263–275.
CrossRef - Hof H. Critical annotations to the use of azole antifungals for plant protection. Antimicrobial agents and chemotherapy 2001;45(11): 2987-2990.
CrossRef - Lo H.J., Tsai S.H., Chu W.L., Chen Y.Z., Zhou Z.L., Chen H.F., Le C.F., Yang Y.L. Fruits as the vehicle of drug-resistant pathogenic yeasts. The Journal of infection2017; 75(3): 254–262.
CrossRef - Devi P.I., ThomasJ.,Raju R.K. Pesticide consumption in India: A spatiotemporal analysis. Agricultural Economics Research Review2017; 30(1): 163-172.
CrossRef - Iatta R., Caggiano G., Cuna T.,Montagna M.T. Antifungal susceptibility testing of a 10-year collection of Candida spp. isolated from patients with candidemia. Journal of chemotherapy (Florence, Italy)2011; 23(2): 92–96.
CrossRef - Warnock D.W. Trends in the epidemiology of invasive fungal infections. Nihon IshinkinGakkaizasshi. Japanese journal of medical mycology2007; 48(1): 1–12.
CrossRef - Yang Y.L., Ho Y.A., Cheng H.H., Ho M., Lo H.J. Susceptibilities of Candida species to amphotericin B and fluconazole: the emergence of fluconazole resistance in Candida tropicalis. Infection control and hospital epidemiology2004; 25(1): 60–64.
CrossRef - Chai L.Y., Denning D.W., Warn P. Candida tropicalis in human disease. Critical reviews in microbiology2010; 36(4): 282–298.
CrossRef - Yang Y.L., Cheng M.F., WangC.W., Wang A.H., Cheng W.T., Lo H.J., Tsary Hospitals. The distribution of species and susceptibility of amphotericin B and fluconazole of yeast pathogens isolated from sterile sites in Taiwan. Medical Mycology2010; 48(2): 328–334.
CrossRef - Yang Y.L., Lin C.C., Chang T.P., Lauderdale T.L., Chen H.T., Lee C.F., Hsieh C.W., Chen P.C., LoH.J. Comparison of human and soil Candida tropicalis isolates with reduced susceptibility to fluconazole. PLOS One 2012;7(4): e34609.
CrossRef - Pfaller M.A., Moet G.J., Messer S.A., Jones R.N.,Castanheira M. Geographic variations in species distribution and echinocandin and azole antifungal resistance rates among Candida bloodstream infection isolates: report from the SENTRY Antimicrobial Surveillance Program (2008 to 2009). Journal of clinical microbiology 2011;49(1): 396–399.
CrossRef - Azevedo M.M., Faria-Ramos I., Cruz L.C., Pina-Vaz C., Goncalves Rodrigues A. Genesis of azole antifungal resistance from agriculture to clinical settings. Journal of agricultural and food chemistry2015; 63(34): 7463-7468.
CrossRef - Arendrup M.C. Update on antifungal resistance in Aspergillus and Candida. Clinical microbiology and infection2014; 20: 42-48.
CrossRef - Snelders E., Rijs A.J., Kema G.H., Melchers W.J., Verweij P.E. Possible environmental origin of resistance of Aspergillusfumigatus to medical triazoles. Environ. Microbiology2009; 75(12): 4053-4057.
CrossRef - Verweij P.E., Snelders E., Kema G.H., Mellado E., Melchers W.J. Azole resistance in Aspergillusfumigatus: a side-effect of environmental fungicide use? The Lancet infectious diseases2009; 9(12). 789-795.
CrossRef - Chowdhary A., Kathuria S., Xu J.,Meis J.F. Emergence of azole-resistant Aspergillusfumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS pathogens2013; 9(11).
CrossRef - Russell P. Fungicide resistance: Occurrence and management. The Journal of Agricultural Science1995; 124(3): 317-323.
CrossRef - Perea S., López-Ribot J.L., Wickes B.L., Kirkpatrick W.R., Dib O.P., Bachmann S.P., Keller S.M., Martinez M. Patterson T.F. Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrobial Agents and Chemotherapy2002; 46(6):1695-703.
CrossRef - Marichal P., Koymans L., Willemsens S., Bellens D., Verhasselt P., Luyten W., Borgers M., Ramaekers F., Odds F.C.,VandenBossche H. Contribution of mutations in the cytochrome P450 14alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology (Reading, England)1999; 145 (Pt 10): 2701–2713.
CrossRef - White T.C., Marr K.A., Bowden R.A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clinical microbiology reviews1998; 11(2): 382–402.
CrossRef - Coleman J.J.,Mylonakis E. Efflux in fungi: la pièce de résistance. PLOS pathogens 2009; 5(6): e1000486.
CrossRef - Robbins N., Caplan T., Cowen L.E. Molecular Evolution of Antifungal Drug Resistance. Annual review of microbiology2017; 71: 753–775.
CrossRef - Revie N.M., Iyer K.R., Robbins N., Cowen L.E. Antifungal drug resistance: evolution, mechanisms and impact. Current opinion in microbiology2018; 45: 70–76.
CrossRef - Rybak J.M., Muñoz J.F., Barker K.S., Parker J.E., Esquivel B.D., Berkow E.L., Lockhart S.R., Gade L., Palmer G.E., White T.C., Kelly S.L., Cuomo C.A., Rogers P.D. Mutations in TAC1B: a Novel Genetic Determinant of Clinical Fluconazole Resistance in Candida auris. mBio2020; 11(3): e00365-20.
CrossRef - Morschhäuser J., Barker K.S., Liu T.T., BlaB-Warmuth J., HomayouniR., Rogers P.D. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoSPathog 2007; 3(11):e164.
CrossRef - Vasicek E.M., Berkow E.L., Flowers S.A., Barker K.S., Rogers P.D. UPC2 is universally essential for azole antifungal resistance in Candida albicans. Eukaryotic cell 2014; 13 (7): 933-946.
CrossRef - Tobudic S., Kratzer C.,Presterl E.Azole‐resistant Candida spp.–emerging pathogens? Mycoses2012; 55: 24-32.
CrossRef - Flowers S.A., Barker K.S., Berkow E.L., TonerG., Chadwick S.G., Gygax S.E., Morschhäuser J., Rogers P.D. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryotic cell2012; 11(10): 1289–1299.
CrossRef - Morio F., Pagniez F., Lacroix C., Miegeville M., Le Pape P. Amino acid substitutions in the Candida albicans sterol Δ5,6-desaturase (Erg3p) confer azole resistance: characterization of two novel mutants with impaired virulence. The Journal of antimicrobial chemotherapy2012; 67(9): 2131–2138.
CrossRef - Hull C.M., Parker J.E., Bader O., Weig M., Gross U., Warrilow A.G., Kelly D.E., Kelly S.L. Facultative sterol uptake in an ergosterol-deficient clinical isolate of Candida glabrataharboring a missense mutation in ERG11 and exhibiting cross-resistance to azoles and amphotericin B. Antimicrobial agents and chemotherapy2012; 56(8): 4223–4232.
CrossRef - Lockhart S.R., Etienne K.A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N.P., ColomboA.L., Calvo B., Cuomo C.A., Desjardins C.A., BerkowE.L., Castanheira M., Magobo R.E., Jabeen K., Asghar R.J., Meis J.F., Jackson B., Chiller T.,Litvintseva A.P. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 2017; 64(2):134–140.
CrossRef - Lee Y., Puumala E., Robbins N., Cowen L.E. Antifungal Drug Resistance: Molecular Mechanisms in Candida albicansand beyond. Chemical reviews2020; 10.1021/acs.chemrev.0c00199. 2020.
CrossRef - Cowen L.E., Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science (New York, N.Y.)2005; 309(5744): 2185–2189.
CrossRef - Kelly S.L., Lamb D.C., Kelly D.E., Manning N.J., Loeffler J., Hebart H., Schumacher U.,Einsele H. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta5,6-desaturation. FEBS Letters1997; 400(1): 80–82.
CrossRef - Spettel K., Barousch W., Makristathis A., Zeller I., Nehr M., Selitsch B., Lackner M., Rath P.M., Steinmann J.,Willinger B. Analysis of antifungal resistance genes in Candida albicans and Candida glabrata using next-generation sequencing. PLOS One2019; 14(1): e0210397.
CrossRef - Kim S.H., Iyer K.R., Pardeshi L., Muñoz J.F., Robbins N., Cuomo C.A., Wong K.H., Cowen L. E. Genetic Analysis of Candida aurisImplicates Hsp90 in Morphogenesis and Azole Tolerance and Cdr1 in Azole Resistance. mBio 2019; 10(1): e02529-18.
CrossRef - Cowen L.E., Steinbach W.J. Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryotic cell2008; 7(5): 747–764.
CrossRef - Selmecki A., Bergmann S., Berman J. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Molecular Microbiology2005; 55(5): 1553–1565.
CrossRef - Selmecki A., Forche A., Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science (New York, N.Y.)2006; 313(5785): 367–370.
CrossRef - Coste A., Selmecki A., Forche A., Diogo D., Bougnoux M.E., d’Enfert C., Berman J.,Sanglard D. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryotic cell2007; 6(10): 1889–1904.
CrossRef - Iwasa M., Makino S., Asakura H., Kobori H., Morimoto Y. Detection of Escherichia coli O157: H7 from Muscadomestica (Diptera: Muscidae) at a cattle farm in Japan. Journal of medical entomology1999; 36(1): 108–112.
CrossRef - Talley J.L., Wayadande A.C., Wasala L.P., Gerry A.C., Fletcher J., DeSilva U., Gilliland S.E. Association of Escherichia coli O157: H7 with filth flies (Muscidae and Calliphoridae) captured in leafy greens fields and experimental transmission of E. coli O157: H7 to spinach leaves by house flies (Diptera: Muscidae). Journal of food protection 2009; 72(7): 1547–1552.
CrossRef - Sela S., Nestel D., Pinto R., Nemny-Lavy E., Bar-Joseph M. Mediterranean fruit fly as a potential vector of bacterial pathogens. Applied and environmental microbiology2005; 71(7): 4052–4056.
CrossRef - Pollack S.Consumer Demand for Fruit and Vegetables: The U.S. Example: Economic Research Service. Washington, DC, USA: United States Department of Agriculture. 2001.
- Sivapalasingam S., Friedman C.R., Cohen L.,Tauxe R.V.Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. Journal of food protection2004; 67(10): 2342–2353.
CrossRef - Berger C.N., Sodha S.V., Shaw R.K., Griffin P.M., Pink D., Hand P., Frankel G. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environmental Microbiology 2010;12(9): 2385–2397.
CrossRef - Mahon B.E., Pönkä A., Hall W.N., Komatsu K., Dietrich S.E., Siitonen A., Cage G., Hayes P.S., Lambert-Fair M.A., Bean N.H., Griffin P.M.,Slutsker L. An international outbreak of Salmonella infections caused by alfalfa sprouts grown from contaminated seeds. The Journal of infectious diseases 1997; 175(4): 876–882.
CrossRef - Denning D.W. Calling upon all public health mycologists: To accompany the country burden papers from 14 countries. European journal of clinical microbiology and infectious diseases: official publication of the European Society of Clinical Microbiology2017; 36(6): 923–924.
CrossRef - Aparicio J.F., Mendes M.V., Antón N., Recio E., Martín J.F.Polyene macrolide antibiotic biosynthesis. Current medicinal chemistry2004; 11(12): 1645–1656.
CrossRef